Ding et al. (GEO accession: GSE290426) studied the transcriptomic profile of wildtype, heterozygous and homozygous ACTA2 (R179H) mutant, and CRISPR/Cas9 ABE-corrected iPSC-derived smooth muscle cells. The ACTA2 gene encodes an actin monomer in vascular smooth muscle cells. The ACTA2 R179H mutation causes multisystemic smooth muscle dysfunction syndrome (MSMDS). To follow the tutorial, please download the raw RNA-seq expression matrix and the metadata files.
Start the analysis of the data by clicking "Start Analysis" after selecting "RNA-Seq Analysis" and "Raw count". In the next tab, upload the expression matrix and metadata files and click "Read data".
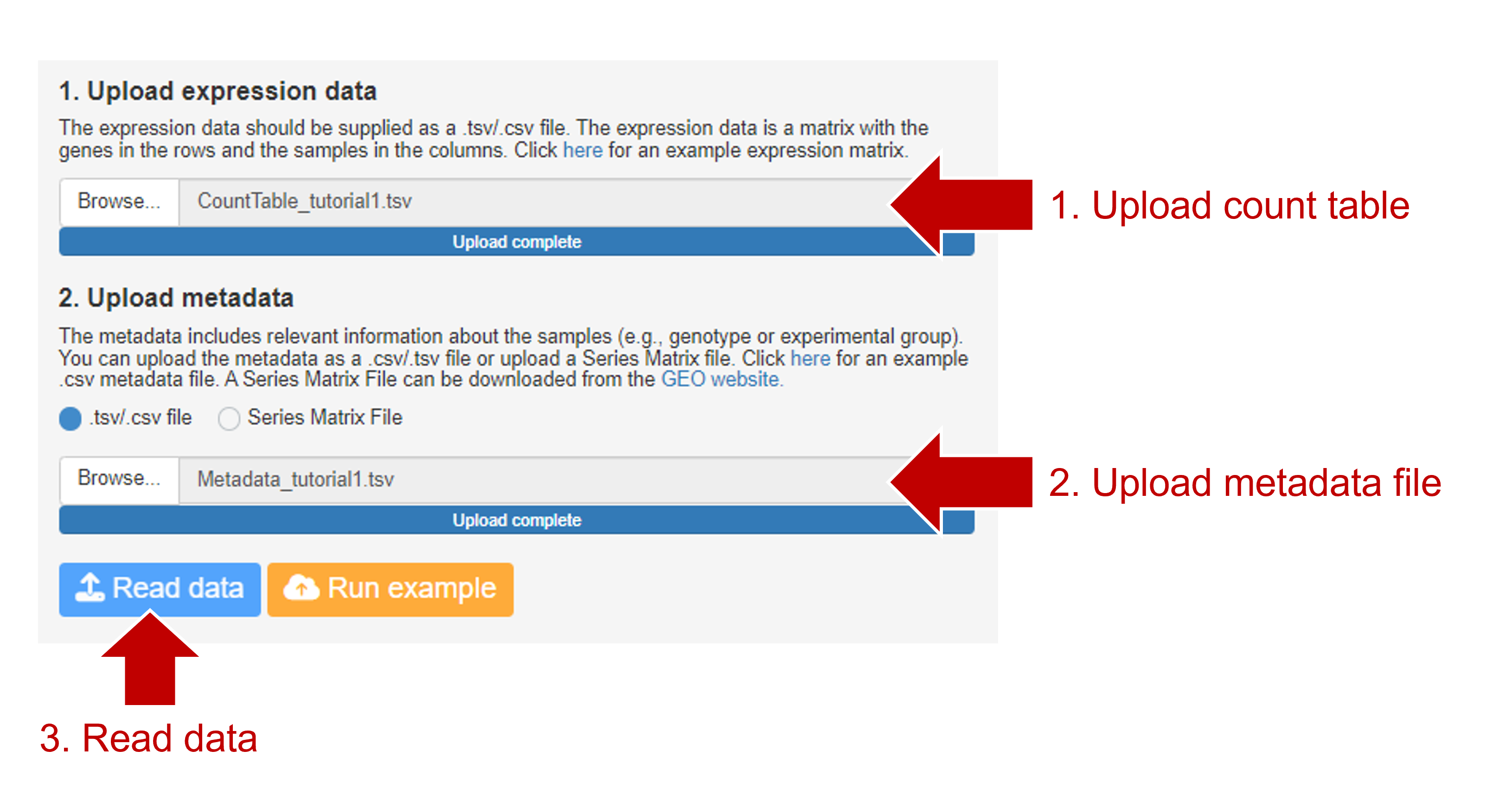
After uploading the data, you can see the metadata and a preview of the count table. In the metadata, you can see that there are four experimental groups: Wildtype (2x wildtype ACTA2 allele), Homozygous (2x mutant ACTA2 allele), Heterozygous (mutant/wildtype ACTA2 allele), and Corrected (ACTA2 mutation corrected by CRISPR/Cas9).
Perform pre-processing with default settings (Remove samples: "keep all samples", Select experimental group: "Group", Filtering: "10").
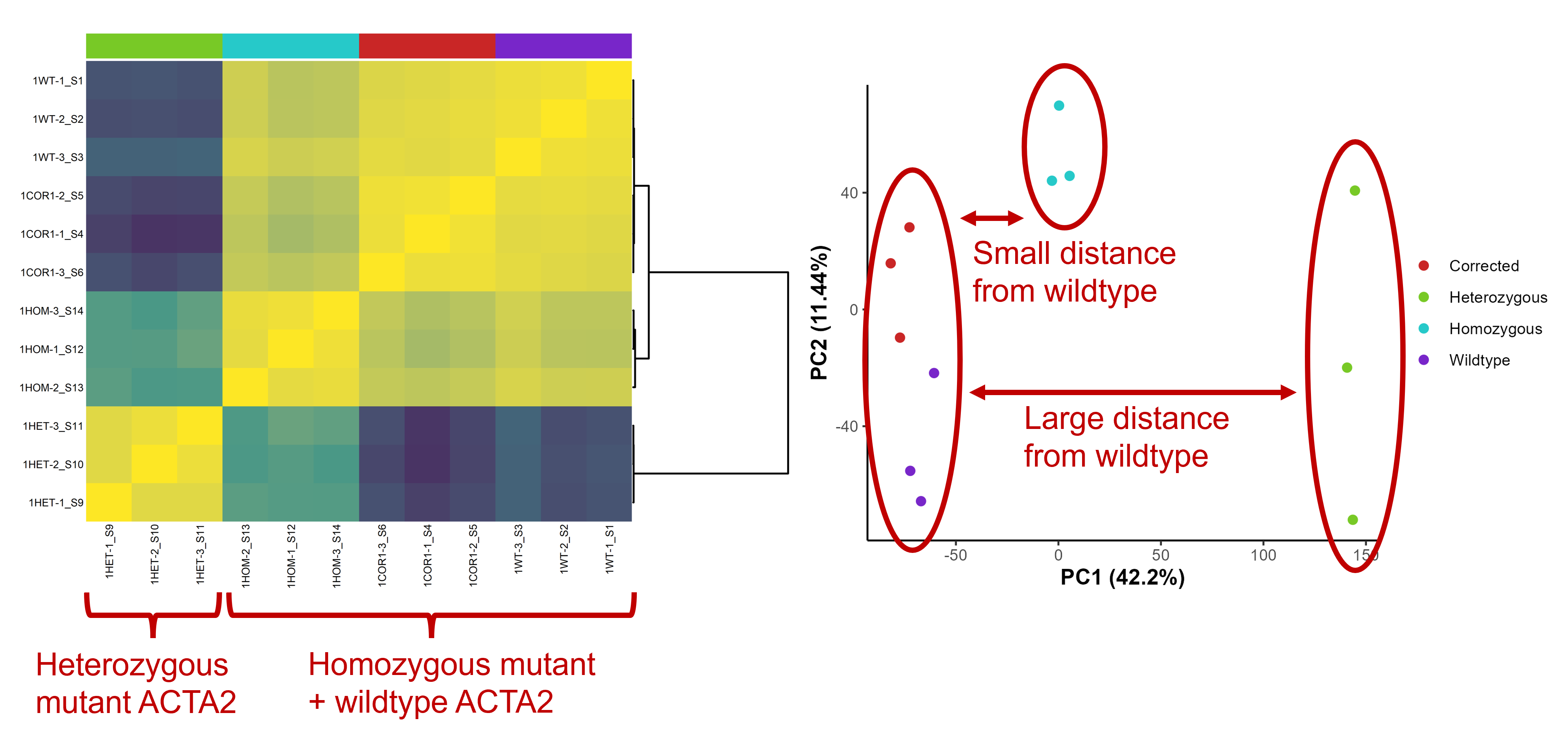
From the QC plots, there does not seem to be any outlier. However, something remarkable can be seen in the sample-sample correlation heatmap and PCA plot. In these plots, you can see that, compared to wildtype samples, heterozygous mutant samples exhibit a more deviating transcriptomic profile than homozygous mutant samples.
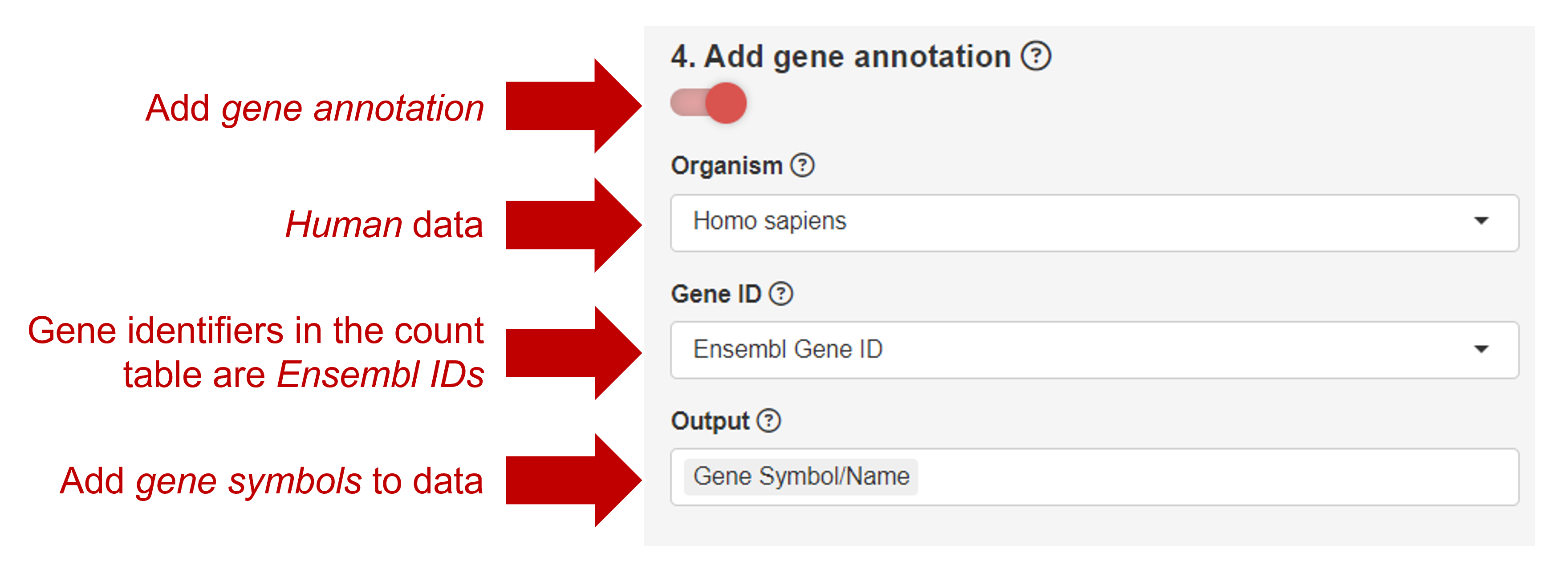
Given the observation in the pre-processing step, we would like to identify genes that are differentially expressed between homozygous mutant and heterozygous mutant. Select the comparison "Heterozygous - Homozygous". Furthermore, add gene symbols to the data, this will aid with the interpretation of the data.
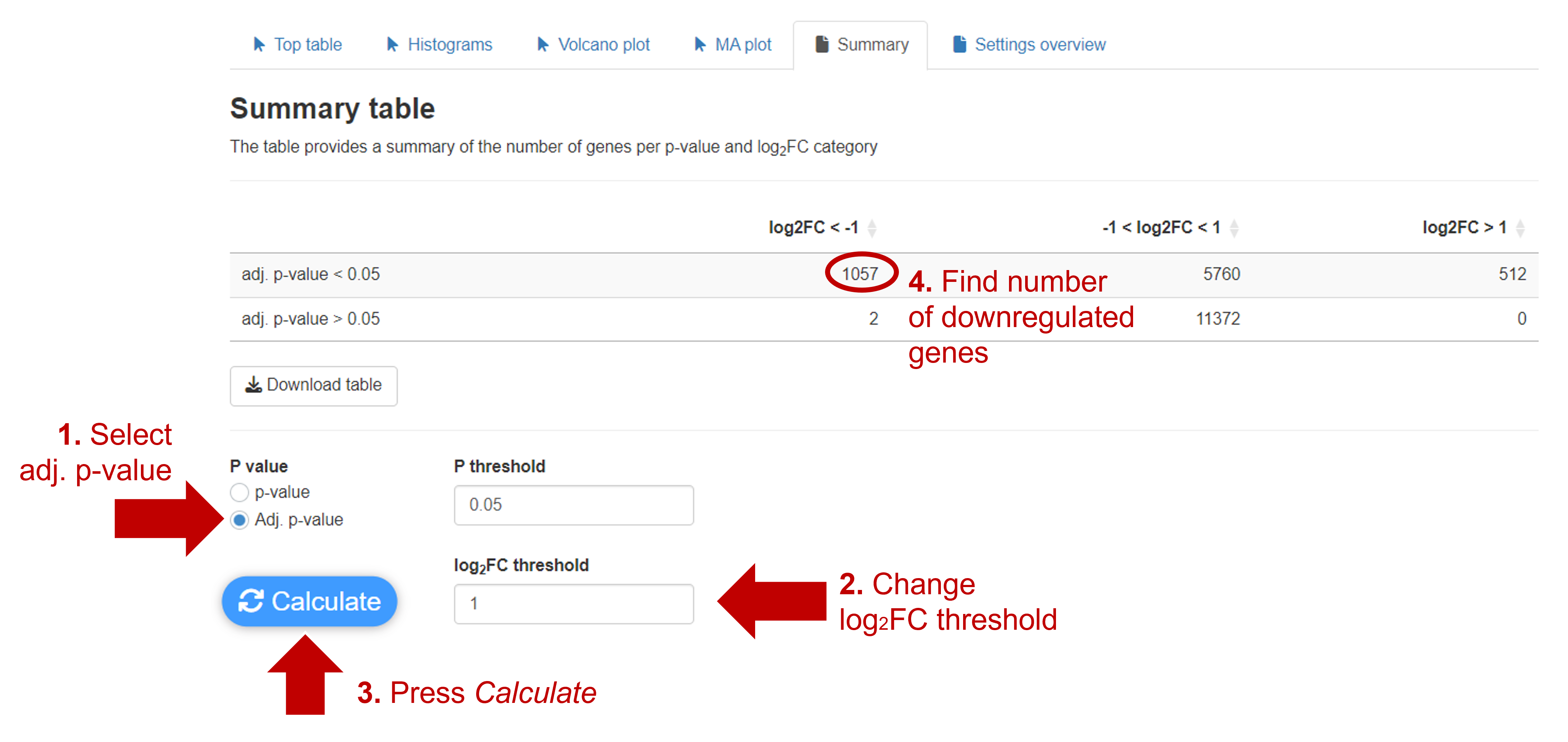
In the Summary tab you can calculate how many genes are up and downregulated for different p-value and log2FC thresholds. Here you can see that 1057 genes are downregulated.
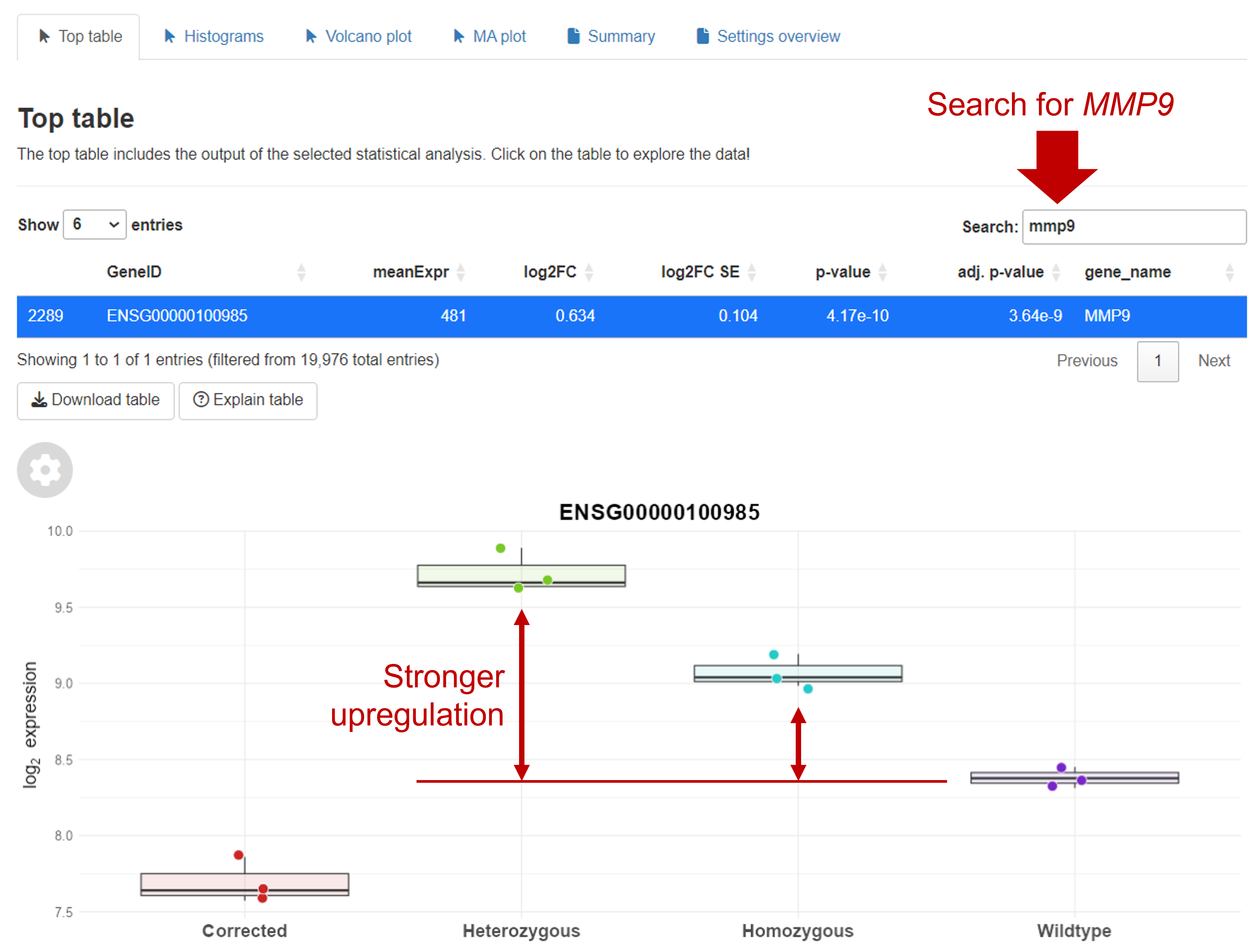
You can look up MMP9 in the Top table. In the boxplots, you can see that MMP9 is stronger upregulated in heterozygous mutant samples.
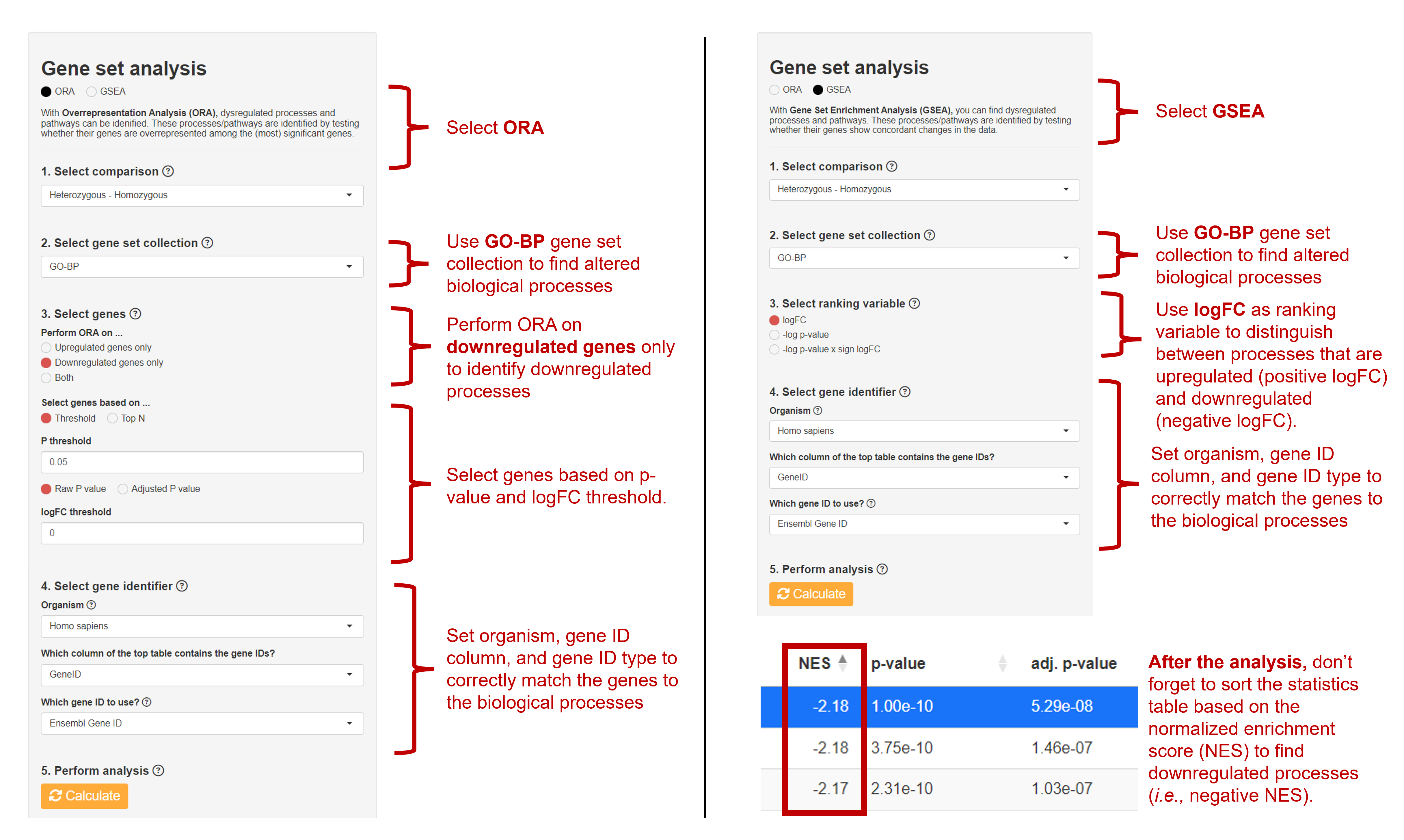
You can now perform gene set analysis to identify which biological processes and pathways are altered.
You can identify downregulated processes by performing overrepresentation analysis (ORA) or Gene Set Enrichment Analysis (GSEA). With both methods, you will find that processes related to cell division, such as chromosome seperation, DNA replication, and mitosis, are downregulated.