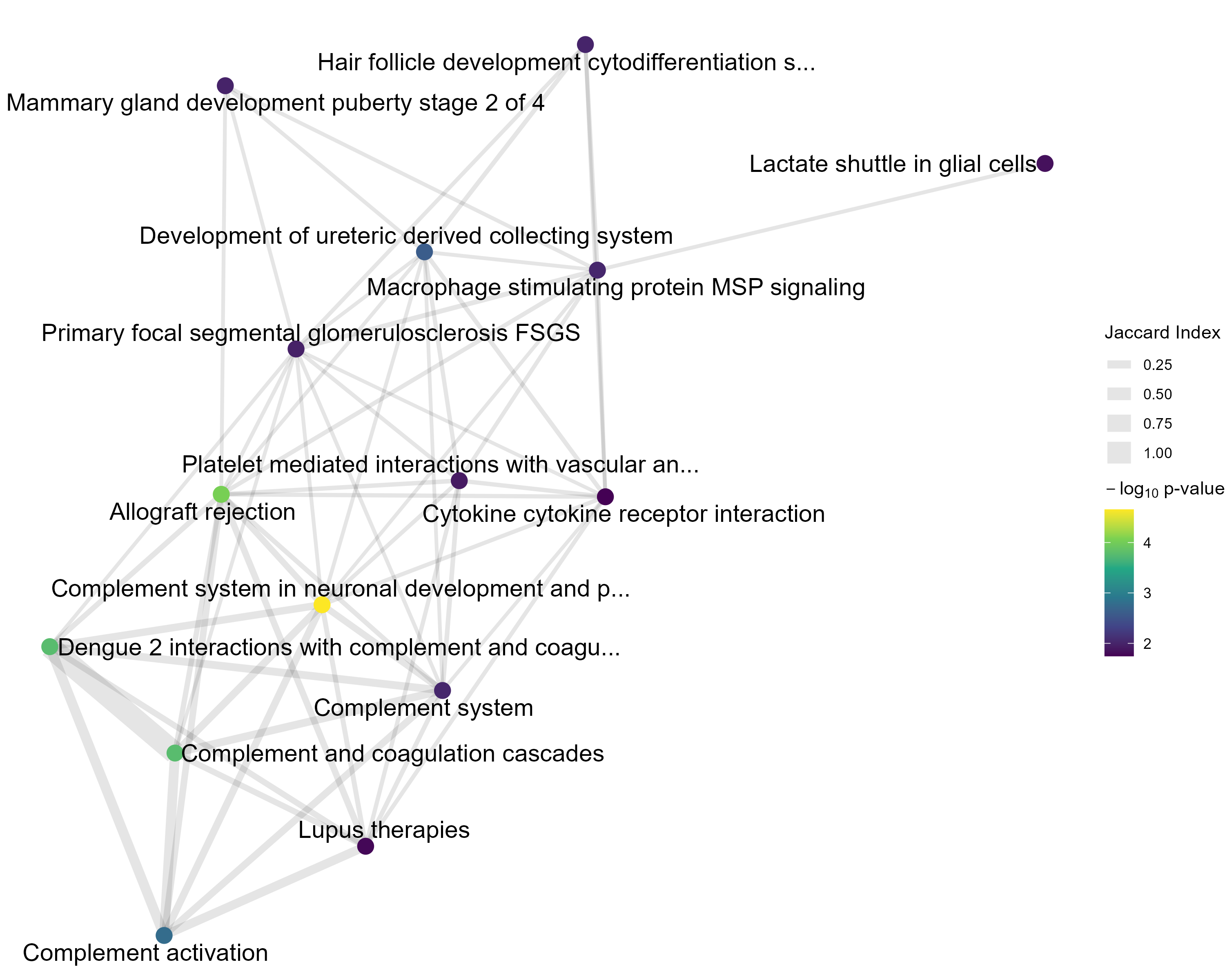
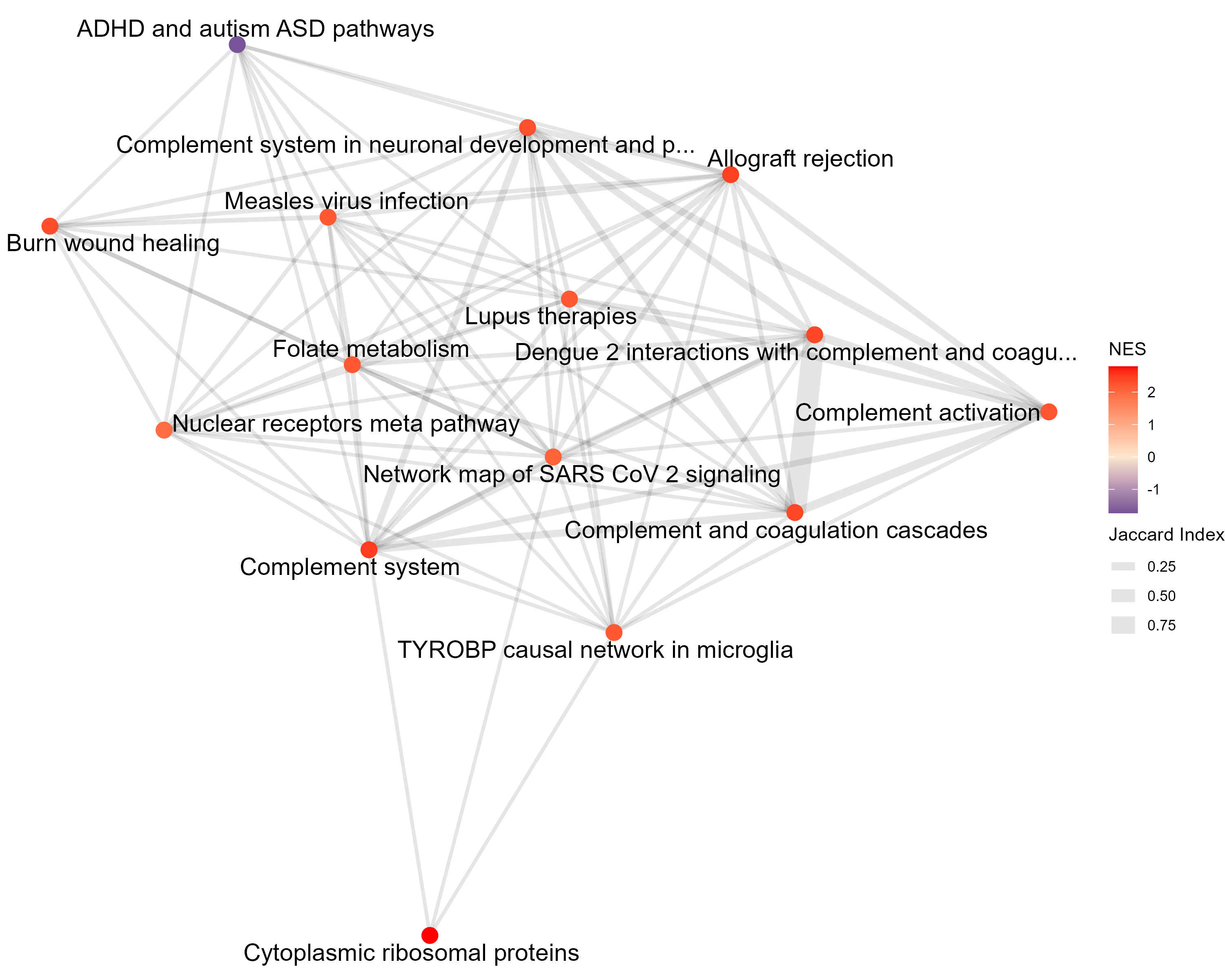
This network visualizes the similarity between the top gene sets identified in the gene set analysis. Each node represents a gene set, and edges indicate similarity between gene sets, quantified by the Jaccard index — defined as the ratio of the number of shared genes (intersection) to the total number of unique genes (union) across the two sets. Higher Jaccard values correspond to greater overlap between gene sets. For Overrepresentation Analysis (ORA) results, nodes are colored based on the number of significant genes contained within each gene set — darker or more intense colors indicate more significant genes. For Gene Set Enrichment Analysis (GSEA) results, nodes are colored by the normalized enrichment score (NES): positive NES values indicate gene sets enriched among upregulated genes, while negative NES values indicate enrichment among downregulated genes. This plot helps you identify clusters of related gene sets and assess whether functionally similar pathways show consistent enrichment patterns.